This is the homepage for the paper
"On Weighting Clustering"
by R. Nock and F. Nielsen, accepted to IEEE Transactions on Pattern Analysis and Machine Intelligence.
Draft
The draft of the paper is available here (1067 Ko) .
.
C++ sources
The gzipped binaries (Linux) are available: click here (34 Ko) .
(34 Ko) .
Unzip the file, move to the subdirectory created, compile using the
makefile, and run (see below for the command line).
Linux binaries
The gzipped binaries (Linux) are available: click here (56 Ko) .
(56 Ko) .
Unzip the file (gzip -d Clustering-Journal-2006-Binaries.tar.gz), move to the subdirectory created,
and then enter a command line.
A slight help is available by entering
Below is this help you get. Please notice that the program also generates images, computed over the two
first coordinates of the points.
Using Clusters :
./cluster.exe [option]* string_name
Options:
h : display this help
d : dimension of the problem (d>=1, default is 2)
n : number of points (default is 1000)
k : number of theoretical clusters (default is 10)
g : type of clusters (0=Gaussian,
1=Ring Gaussian,
2=Grid Gaussian, BIRCH-like, with dimension==2,
3=0 with overlap reduced,
4=Hypercubic uniform, default is 0)
b : balance between sampling the theoretical clusters (0=Uniform: all proportions are identical,
1=Skewed random: each proportion is picked at random,
2=Unbalanced: proportions are approximately exponentially decreasing, default is 0)
K : number of experimental clusters (default is 5)
I : type of initialisation (0=Random, 1=Forgy, default is 1)
S : number of running steps (default is 10)
N : #steps between saving each image (default is 1,
i.e. each config image is saved)
T : width(=length) of .tif images saving the configs (default is 512)
W : show the weights on the images (0/1, default is 0=no)
C : show the 2D-convex hulls on the images (0/1, default is 0=no)
(please pick 1 only with K Means)
Z : type of algorithm (0=Kmeans, 1=Harmonic Means, 2=EM, 3=Fuzzy C Means, default is Kmeans)
D : diagonal covariance matrices for Gaussian EM (0=No, 1=Yes, default is 1)
--- string_name is a generic name used to save (i) the config images
(by appending [string_name]_image_[#step].tif), (ii) the KMN-Losses
(the filename is [string_name]_params.txt), and (iii) the theoretical
configuration image (the filename is [string_name]_theorique.tif)
the default value for string_name="default"
-Example (cut and paste):
./cluster.exe -d 2 -n 2000 -k 2 -g 1 -K 16 -b 0 -I 0 -S 50 -N 1 -T 200 -W 1 -C 0 -Z 0 my_test
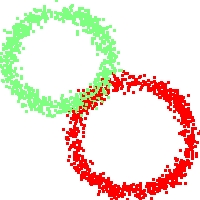
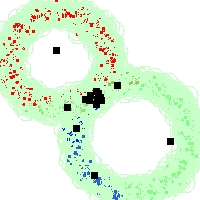
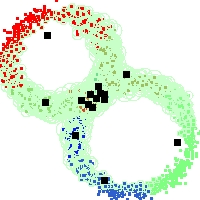
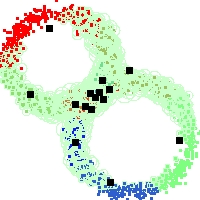
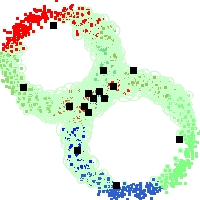
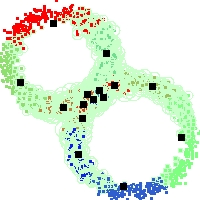
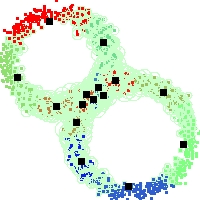
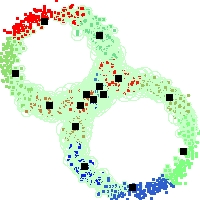
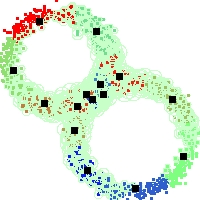
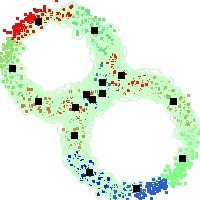
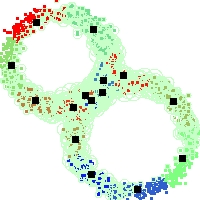
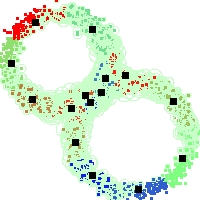
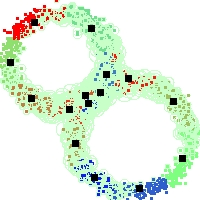
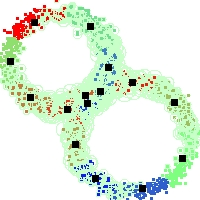
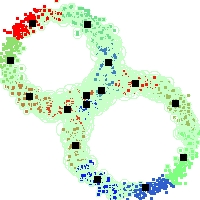
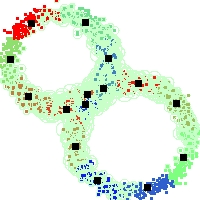
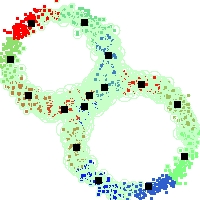
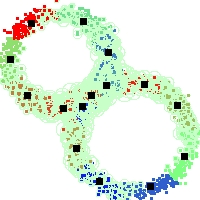
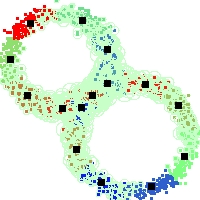
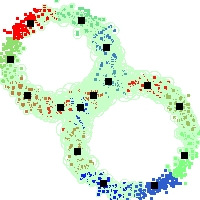
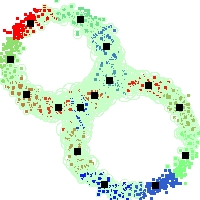
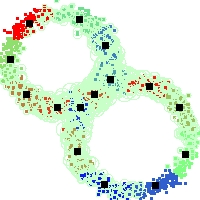
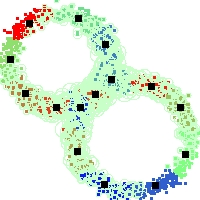
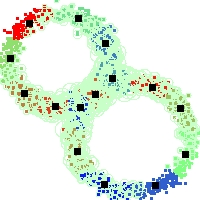
Example of run
Below, you will find the first images output when entering the command line
./cluster.exe -d 2 -n 2000 -k 2 -g 1 -K 16 -b 0 -I 0 -S 50 -N 1 -T 200 -W 1 -C 0 -Z 0 my_test
Some of these images appear in the paper. Notice that the left image presents the theoretical configuration.
Post-processing output files
You will notice that the program outputs a number of statistics, results and files. We have
made some post-processing files to crunch the outputs and deliver some useful statistics
that are used in the paper. You can retrieve them by moving to subdirectory
created when unzipping the .tarfile for the binaries above. Consult the
file on directory Clustering-Journal-2006-Binaries to see the purpose of each post-processing
file. Consult the paper for the meaning of the statistical tests used.
Feedback
Your feedback is much appreciated. Your can reach us at rnock@martinique.univ-ag.fr, or at Nielsen@csl.sony.co.jp.