This is the homepage for the paper
"Grouping with Bias Revisited"
by R. Nock and F. Nielsen, appearing in CVPR'04 (IEEE International Conference on Computer Vision and
Pattern Recognition).
Paper's draft
A draft of the paper is available by clicking here (.pdf).
(.pdf).
Source C
The zipped source C is available, including the flower example below: click here (<400 Ko) .
(<400 Ko) .
Linux binaries and howtos (old)
(these binaries are a bit old now; you may directly use the Source
code above)
The gzipped binary is available: click here (48,503 Ko).
(48,503 Ko).
Running SRM.B is quite easy: simply unzip the file is necessary (gzip -d psis.gz), and then run it !
SRM.B runs on the command line, as for example:
./srmB -B 1 -M 2 -s stpierre.tif stpierre-srm-with-bias.tif
There is a very, very slight help, which you can obtain by running:
Below is this help you get:
Welcome.
srmB is (c) 2003/04 R. Nock, F. Nielsen. It was compiled and run under SuSE Linux.
srmB implements our paper 'Grouping with Bias Revisited', CVPR'04.
This program is provided AS IS, without any warranty. Use it at your own risk !
Usage :
./srmB [option]* -s input.tiff output.tiff
Options:
h : display this help
S : 0,1,2,3 type of output (0=Bordered regions with original pixels,
1=Black-bordered regions with white pixels,
2=Bordered regions with average-color pixels)
3=Randomized colored regions)
B : 0-1, border color (0=black, 1=white, valid for S=0,2)
P : 0-1, saves (when P=1) the (at most) 15 largest regions of the segmentation
in files named fileRegionX, for X=0, 1, ...
b: is models >0, Boolean (0=No/1=Yes) stating whether the bias is to be shown on the output
M: Models specified (0 is no model)
The models have to be stored into separate files named input.tiff_modeleX, for X=1, 2, ...
Default settings: S=B=P=M=0, b=1
Format of model files:
each file contains as rows the coordinates (rom, column) of each pixel model
each file ends by the row -1[tab]-1
example for model file flower.tif_modele1:
195 245
360 320
290 60
-1 -1
--- Comments and suggestions much welcomed, at: rnock@martinique.univ-ag.fr
The program runs using the same parameters as in the paper's experiments.
We refer the reader to the CVPR paper.
The experiments of this webpage displays some experiments, and the tiff source images of which are available.
Note that psis is implemented to work on tiff RGB (color 8-bits) images.
Images and experiments
The following tables display the additional result of the algorithm SRM.B on various images.
Please note the following:
- images are represented with their original size;
- for each image, we also give some of the largest regions found by SRM.B;
- the image names are those of the drafts; if you click on the image name, you retrieve a gzipped .tif copy of the original;
- for each image segmented with bias, we give the bias files used;
- to avoid eventual downloading problems for the images, we suggest to right click + select "Save Link As..." on the desired image link;
- command lines are also given: cut and paste to rerun the experiments.
Flower (Fig. 1)
(Fig. 1)
Command line :
./srmB -S 0 -B 1 -P 1 -b 0 -M 3 -s flower.tif flower-srm-with-bias.tif
Models :
- flower.tif_modele1 : click here
 ,
,
- flower.tif_modele2 : click here
 ,
,
- flower.tif_modele3 : click here
 .
.
| Original image |
 |
| SRM.B |
 |
castle (new17) (Fig. 2)
(Fig. 2)
Command line :
./srmB -S 0 -B 1 -P 1 -s new17.tif new17-srm-without-bias.tif
| Original image |
 |
| SRM.B |
 |
| SRM.B #1 | SRM.B #2 | SRM.B #3 | SRM.B #4 | SRM.B #5 | SRM.B #6 |
SRM.B #7 |
 |
 |
 |
 |
 |
 |
 |
Saint-Pierre (stpierre) (Fig. 2)
(Fig. 2)
Command line :
./srmB -S 0 -B 1 -P 1 -s stpierre.tif stpierre-srm-without-bias.tif
| Original image |
 |
| SRM.B |
 |
| SRM.B #1 | SRM.B #2 | SRM.B #3 | SRM.B #4 | SRM.B #5 | SRM.B #6 |
SRM.B #7 |
 |
 |
 |
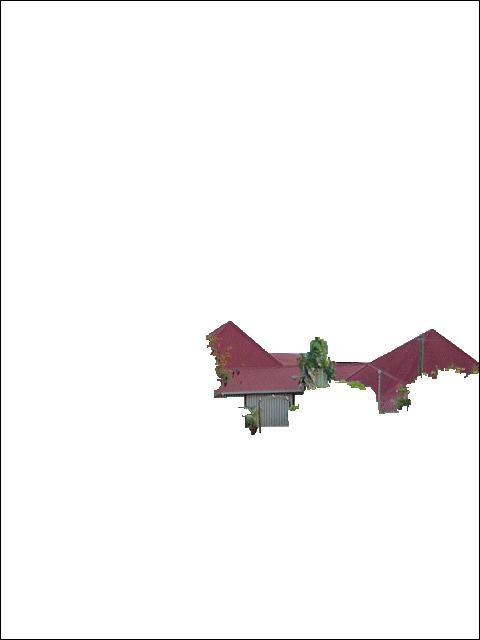 |
 |
 |
 |
leopard (Fig. 3)
(Fig. 3)
Command line :
./srmB -S 0 -B 1 -P 1 -M 2 -s leopard.tif leopard-srm-with-bias.tif
Models :
- leopard.tif_modele1 : click here
 ,
,
- leopard.tif_modele2 : click here
 .
.
| Original image |
 |
| SRM.B |
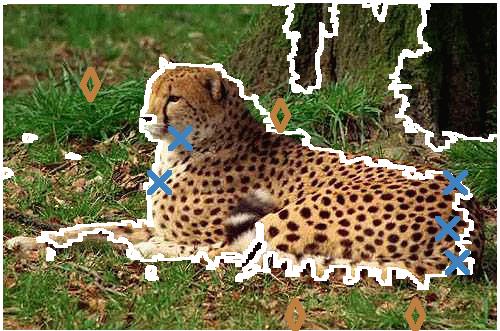 |
| SRM.B #1 | SRM.B #2 | SRM.B #3 |
 |
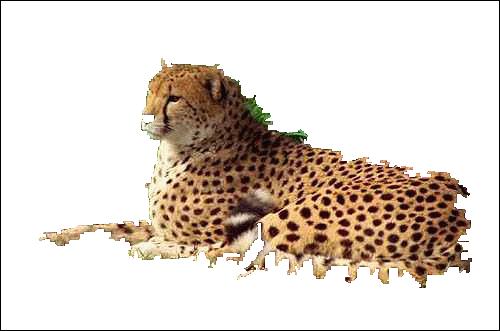 |
 |
badger (blaireau) (Fig. 3)
(Fig. 3)
Command line :
./srmB -S 0 -B 1 -P 1 -M 2 -s blaireau.tif blaireau-srm-with-bias.tif
Models :
- blaireau.tif_modele1 : click here
 ,
,
- blaireau.tif_modele2 : click here
 .
.
| Original image |
 |
| SRM.B |
 |
| SRM.B #1 | SRM.B #2 |
 |
 |
flower (Fig. 4)
(Fig. 4)
Command line :
./srmB -S 0 -B 1 -P 1 -M 3 -s flower.tif flower-srm-with-bias.tif
Models :
- flower.tif_modele1 : click here
 ,
,
- flower.tif_modele2 : click here
 ,
,
- flower.tif_modele3 : click here
 .
.
| Original image |
 |
| SRM.B |
 |
| SRM.B #1 | SRM.B #2 | SRM.B #3 |
 |
 |
 |
new09 (castle2)  (Fig. 5)
(Fig. 5)
Command line :
./srmB -S 0 -B 1 -P 1 -M 3 -s new09.tif new09-srm-with-bias.tif
Models :
- new09.tif_modele1 : click here
 ,
,
- new09.tif_modele2 : click here
 ,
,
- new09.tif_modele3 : click here
 .
.
| Original image |
 |
| SRM.B |
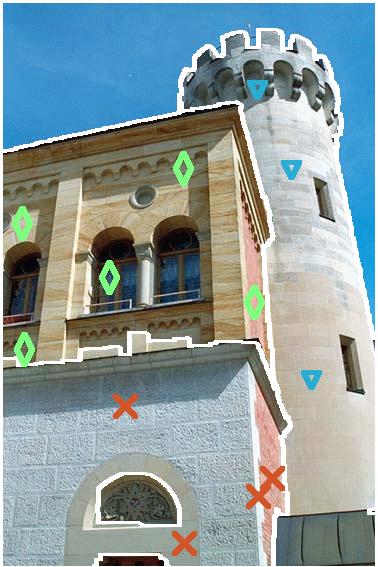 |
| SRM.B #1 | SRM.B #2 | SRM.B #3 | SRM.B #4 |
 |
 |
 |
 |
BSDB experiments
Additional experiments of SRM.B (and other
image processing algorithms as well) are available on The Berkeley Segmentation Dataset and Benchmark. A webpage
is devoted to this repository, and you will find it here .
.
Windows material
To get the Windows version of SRM.B, please click here .
.
Feedback
Your feedback is much appreciated. Your can reach us at rnock@martinique.univ-ag.fr,
or at Nielsen@csl.sony.co.jp.







































